3. Bioschemas with Mkdocs
Introduction
In this example, we use the ELIXIR Lesson template since it uses mkdocs as static site generator and comes with a Python library addbioschemas which makes adding Bioschemas markup quite easy. For the ELIXIR Lesson template, an extensive tutorial has been published which describes Bioschemas markup using addbioschemas.
Bioschemas profiles for training resources
Many training related resources will include pages describing tutorials or courses. As such, they are marked up using the following three profiles:
TrainingMaterial: A profile describing training materials in life sciences, it can be used on its own (as it happens with the Bioschemas tutorials) or in combination with aCourseInstance.Course: A profile describing a course from a generic point of view, i.e., the learning objectives of a course rather than where and when it is delivered.CourseInstance: A profile describing a particular instance of a course, i.e., an edition of a course that is scheduled for specific dates and happening in a specific location (that of course can be online or on-site, virtual or real).
Note that the CourseInstance profile is used in tandem with the Course profile, i.e., a CourseInstance does not exist without a Course but a Course can exist without a CourseInstance (there are no current offerings of the course).
Setup of the repository
Let’s start with the first step and create a new repository from the Bioschemas tutorial template for mkdocs. Once you have opened the link in your browser, you can create a new GitHub repository by clicking on the green ‘Use template’ button. Enter a new name for the created repository e.g. ELIXIR-TrP-Bioschemas-mkdocs under your own GitHub account.
Markup for a training course page using mkdocs as static site generator
We reuse the technical instruction given in the ELIXIR Lesson template tutorial to add markup using the libray addbioschemas. But instead of using an annotation according to Bioschemas profile LearningResource we start with the profiles Course/CourseInstance.
Open the file _data/metadata.yml and add the following:
"@context": https://schema.org/
"@type": Course
"@id": https://elixir-europe-training.github.io/ELIXIR-TrP-LessonTemplate-MkDocs/
http://purl.org/dc/terms/conformsTo:
"@type": CreativeWork
"@id": https://bioschemas.org/profiles/Course/1.0-RELEASE
description: ELIXIR Bioschemas Annotation Guide
keywords: Bioschemas, Schema validation, Harvesting markup, Deploying markup
name: Bioschemas Annotation Guide for training material
# lookup at https://spdx.org/licenses/
license: CC-BY-4.0
educationalLevel: beginner
teaches: [
"The student will be able to recall shell commands",
"The student will be able to write code to copy files",
"The student will be able to discover new commands on their own"
]
hasCourseInstance:
- "@type": CourseInstance
courseMode: online
location: Virtual
startDate: 2022-01-10
endDate: 2022-01-10
inLanguage: en-GB
url: https://www.workshops.org
http://purl.org/dc/terms/conformsTo:
- "@type": CreativeWork
"@id": https://bioschemas.org/profiles/CourseInstance/1.0-RELEASE
instructor:
- "@type": Person
name: Elin Kronander
identifier: https://orcid.org/0000-0003-0280-6318
The rather elaborate annotation according to Bioschemas profiles Course/CourseInstance which will be created by the bioschemas library looks like this.
{
"@context":"https://schema.org/",
"@type":"Course",
"http://purl.org/dc/terms/conformsTo":{
"@type":"CreativeWork",
"@id":"https://bioschemas.org/profiles/Course/1.0-RELEASE"
},
"description":"Bioschemas Tutorial at SWAT4HCLS Leiden",
"keywords":"Bioschemas, SWAT4HCLS, Schema validation, Harvesting markup, Deploying markup",
"name":"Bioschemas - Deploying and Harvesting Markup",
"hasCourseInstance":[
{
"@type":"CourseInstance",
"http://purl.org/dc/terms/conformsTo":{
"@type":"CreativeWork",
"@id":"https://bioschemas.org/profiles/CourseInstance/0.8-DRAFT-2020_10_06"
},
"courseMode":"online",
"location":"Virtual, and Fletcher Wellness-Hotel, Leiden, The Netherlands",
"startDate":"2022-01-10",
"endDate":"2022-01-10",
"inLanguage":"en",
"url":"https://www.swat4ls.org/workshops/leiden2022/",
"instructor":[
{
"@type":"Person",
"name":"Alban Gaignard",
"@id":"https://bioschemas.org/people/AlbanGaignard",
"url":"https://bioschemas.org/people/AlbanGaignard"
},{
"@type":"Person",
"name":"Leyla Jael Castro",
"@id":"https://bioschemas.org/people/LeylaGarcia",
"url":"https://bioschemas.org/people/LeylaGarcia"
},{
"@type":"Person",
"name":"Alasdair Gray",
"@id":"https://bioschemas.org/people/AlasdairGray",
"url":"https://bioschemas.org/people/AlasdairGray"
}
]
}
]
}
Markup for a training material page using mkdocs as static site generator
We reuse the technical instruction given in the ELIXIR Lesson template tutorial to add markup using the libray addbioschemas.
In this example, we use an annotation according to Bioschemas profile LearningResource.
We will use a yaml file to specify the markup using the Bioschemas profile. In order to have the tutorial marked up with the Bioschemas annotation, it is important to add a specific instruction to the markdown file which contains the initial version of the tutorial file.
In combination with mkdocs-material, this specific instruction should be placed after markdown text, so never at the very top of the page.
## 1.1 First subtopic
[add-bioschemas file='_data/training-material.yml']
Here you can enter text and create inline citations[^Garcia2020] by using the bibtex plugin. Add your references in `references.bib`, and cite [^hoebelheinrich_nancy_j_2022_6769695] by adding the @refid inside brackets like this ``
...
Create a file _data/training-material.yml and add the following yaml:
"@context": https://schema.org/
"@type": LearningResource
"@id": https://elixir-europe-training.github.io/ELIXIR-TrP-LessonTemplate-MkDocs/
http://purl.org/dc/terms/conformsTo:
"@type": CreativeWork
"@id": https://bioschemas.org/profiles/TrainingMaterial/1.0-RELEASE
description: ELIXIR Bioschemas Annotation Guide
keywords: Bioschemas, Schema validation, Harvesting markup, Deploying markup
name: Bioschemas Annotation Guide for training material
# lookup at https://spdx.org/licenses/
license: CC-BY-4.0
inLanguage: en-GB
educationalLevel: beginner
teaches: [
"The student will be able to list the main Bioschemas profiles",
"The student will be able to write Bioschemas markup"
]
version: 2.0
author:
- "@type": Person
name: Niall Beard
identifier: https://orcid.org/0000-0002-2627-0231
url: https://orcid.org/0000-0002-2627-0231
This YAML file is processed by the bioschemas library to embed the following JSON-LD within the created tutorial web page. The rather elaborate annotation according to Bioschemas profile TrainingMaterial looks like this:
{
"@context":"https://schema.org/",
"@type":"LearningResource",
"@id": "https://example.com/training-material/12345",
"dct:conformsTo":{
"@type":"CreativeWork",
"@id":"https://bioschemas.org/profiles/TrainingMaterial/1.0-RELEASE"
},
"description":"This guide will show you how to do add Schema.org markup to a GitHub Pages site.",
"keywords":"schema.org, TeSS, GitHub pages",
"name":"Adding Schema.org to a GitHub Pages site",
"about":,
"author":[
{
"@type":"Person",
"name":"Niall Beard",
"@id":"https://orcid.org/0000-0002-2627-0231",
"url":"https://bioschemas.org/people/NiallBeard"
}
],
"educationalLevel":"beginner",
"identifier": "http://dx.doi.org/12349302",
"inLanguage": "en-UK",
"teaches": [
"The student will be able to list the main Bioschemas profiles",
"The student will be able to write Bioschemas markup"
],
"dateModified":"2021-07-22",
"license":"CC-BY 4.0",
"version":2.0
}
This examples uses most of the properties specified in the Bioschemas profile TrainingMaterial.
Once you have inserted the special instruction, save the markdown file file.
Publish training course and material via GitHub
Start the publishing process by following these instructions.
TODO: add specific instructions for this forked template.
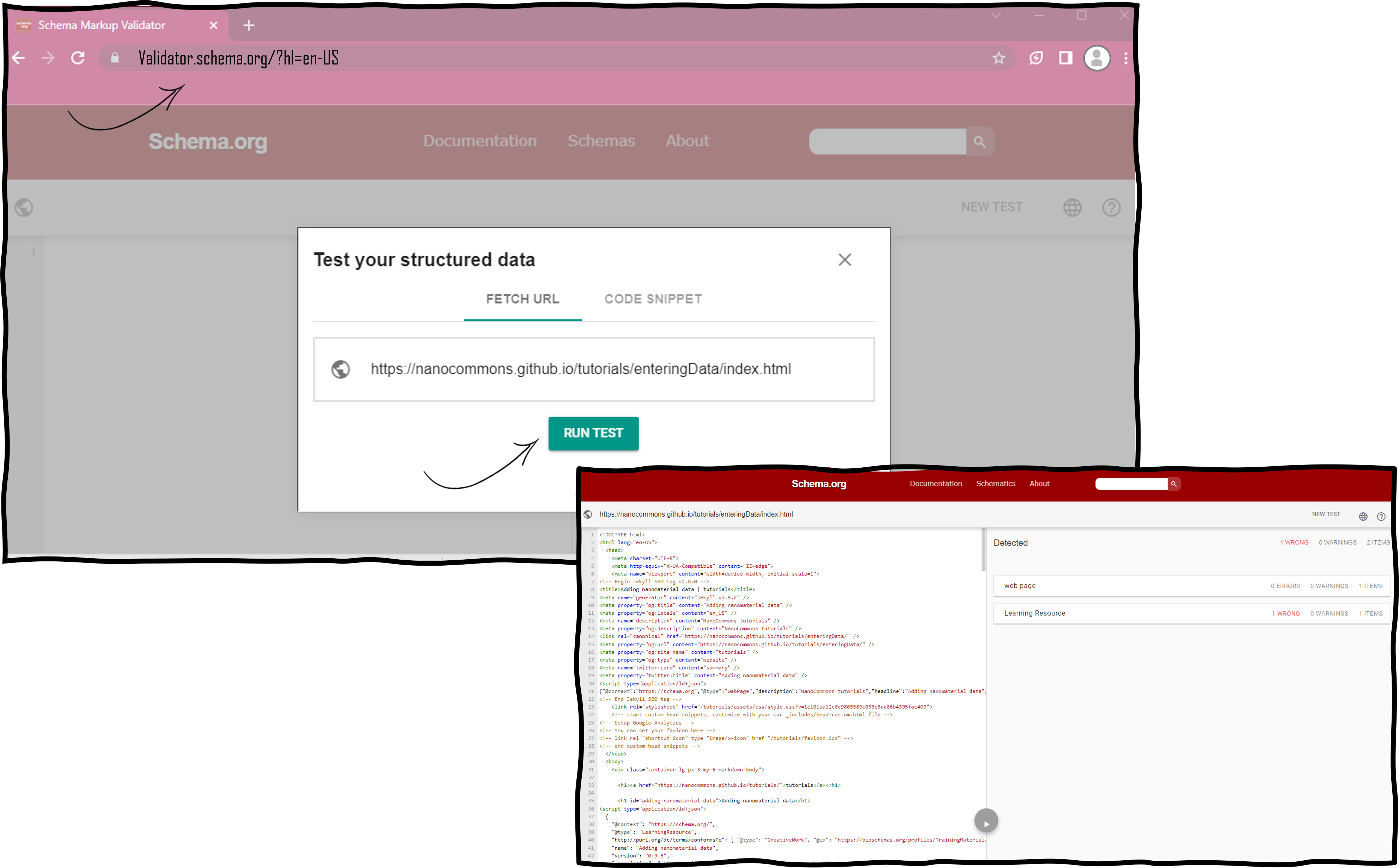
Validate the markup of the page with the Schema.org validator
Validate the individual page with the schema.org validator by pasting the URL into the Fetch URL tab. The validation procedure will indicate if you have used non-existing properties of the Bioschemas profile. If error messages are returned, have a look at the troubleshooting section below.
In this scenario, you can validate the course web site (TODO: add link) and individual tutorial web site (TODO: add link).
-
Leyla Garcia, Bérénice Batut, Melissa L. Burke, Mateusz Kuzak, Fotis Psomopoulos, Ricardo Arcila, Teresa K. Attwood, Niall Beard, Denise Carvalho-Silva, Alexandros C. Dimopoulos, Victoria Dominguez Del Angel, Michel Dumontier, Kim T. Gurwitz, Roland Krause, Peter McQuilton, Loredana Le Pera, Sarah L. Morgan, Päivi Rauste, Allegra Via, Pascal Kahlem, Gabriella Rustici, Celia W.G. Van Gelder, and Patricia M. Palagi. Ten simple rules for making training materials FAIR. PLoS Computational Biology, 16(5):1–9, 2020. doi:10.1371/journal.pcbi.1007854. ↩
-
Nancy J Hoebelheinrich, Katarzyna Biernacka, Michelle Brazas, Leyla Jael Castro, Nicola Fiore, Margareta Hellstrom, Emma Lazzeri, Ellen Leenarts, Paula Maria Martinez Lavanchy, Elizabeth Newbold, Amy Nurnberger, Esther Plomp, Lucia Vaira, Celia W G van Gelder, and Angus Whyte. Recommendations for a minimal metadata set to aid harmonised discovery of learning resources. June 2022. URL: https://doi.org/10.15497/RDA00073, doi:10.15497/RDA00073. ↩